MTM home
XCR
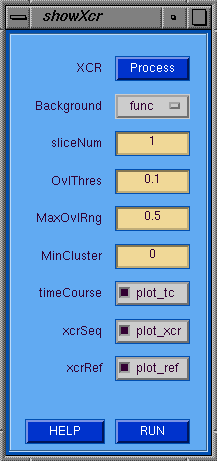
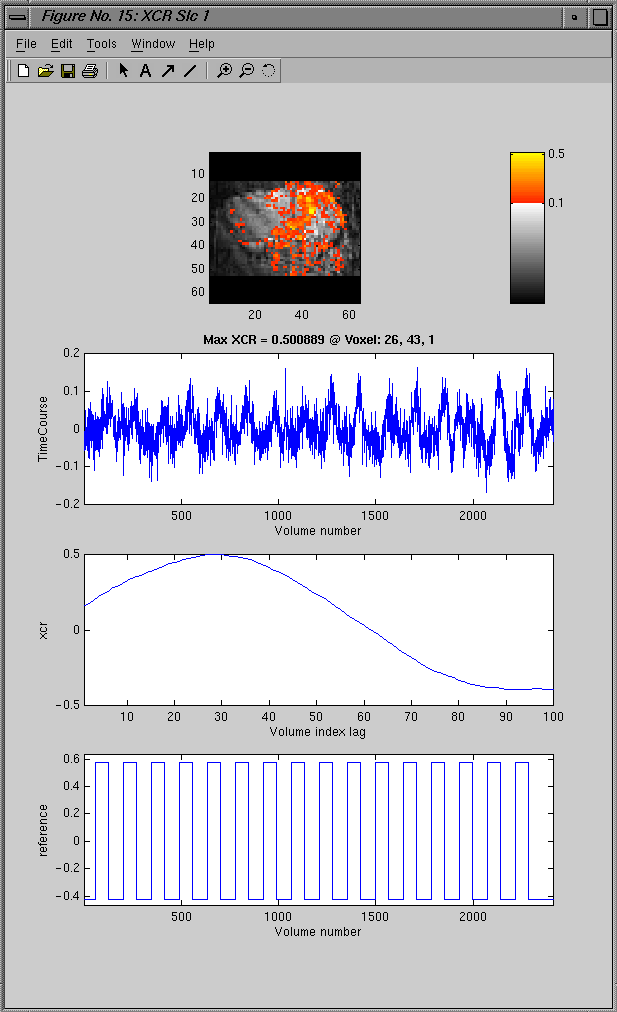
The showXcr GUI allows the user display the results
(i.e. Value of or lag at the maximum correlation)
of a cross correlation as an overlay map on top of the
corresponding slice of either the functional or anatomic
data. The overlay map is color coded along a scale
from the MinOvlRng and MaxOvlRng for those pixels with a
correlation value that is no less than the xcr_thres value
and who belong to a cluster with a size no smaller than
the MinCluster. If the timeCourse plot option is selected,
then when the image is clcked by the mouse the underlying
time series is plotted with optionally the correlation
sequence and/or the reference vector. First the process
button must be pressed to popup the xcr window where the
xcr can be invoked.
showXcr GUI Tips
- XCR --> Popup the Cross-Correlation processing window.
- Background --> Type of image for background.
- sliceNum --> Slice number to display.
- Overlay --> Type of image for overlay.
- xcr_thres --> XCR threshold value for overlay mask.
- MinOvlRng --> Minimun display value for scaling overlay.
- MaxOvlRng --> Maximun display value for scaling overlay.
- MinCluster --> Minimum cluster size for overlay display.
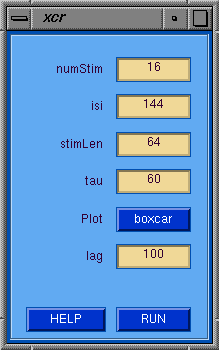
The xcr GUI allows the user to specify the xcr parameters
and invoke the calculation by pressing the RUN button.
xcr GUI Tips
- numStim --> Number of stimulation/task periods.
- isi --> Inter-stimulus interval in volAcq units.
- stimLen --> Stimulation length in volAcq units.
- tau --> Delay until the first stimulation period in volAcq units.
- Plot --> Plot the xcr boxcar reference vector.
- lag --> Number of lags to compute in volAcq units.
Relative URL:/~strupp/mtm_doc/xcr.shtml
Last modified: Thursday, 03-Jan-2002 14:40:19 CST