MATLAB Examples 3 (covering Statistics Lectures 5 and 6)
Contents
Example 1: Demonstration of various types of resampling
% define a set of data data = randn(1,100); % 100 data points n = length(data); % the number of data points % in bootstrapping, we draw n points with replacement from a % set of n data points. numboots = 1000; % how many bootstraps to perform for p=1:numboots bootix = ceil(n*rand(1,n)); % indices to use for the bootstrap sample % data(bootix) gives the bootstrap sample. end % in leave-one-out cross-validation, we leave out a data point, % train on the remaining data points and then test on the left-out % data point. this process is repeated for each data point. for p=1:n trainix = setdiff(1:n,p); % indices to use for training testix = p; % indices to use for testing % data(trainix) gives the training data % data(testix) gives the testing data end % in k-fold cross-validation, we divide the data into k parts, % leave out one of the parts, train on the remaining parts, % and then test on the left-out part. this process is repeated % for each part. k = 8; % how many parts to use allix = randperm(n); % all data indices, randomly ordered numineach = ceil(n/k); % at least one part must have this many data points % add NaNs and reshape data indices into a 2D matrix. % this process ensures that the numbers of data points in each part % are as evenly balanced as possible. allix = reshape([allix NaN(1,k*numineach-n)],k,numineach); for p=1:k testix = allix(p,:); % indices to use for testing testix(isnan(testix)) = []; % remove NaNs if necessary trainix = setdiff(1:n,testix); % indices to use for training % data(trainix) gives the training data % data(testix) gives the testing data end
Example 2: Bootstrap a simple linear model
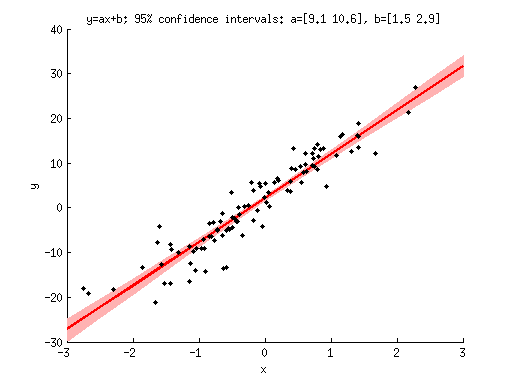
% generate some data n = 100; x = randn(1,n); y = 10*x + 2 + 4*randn(1,n); % define numboots = 10000; % number of bootstraps to perform xvals = -3:.5:3; % x-values to evaluate the model at % perform the bootstraps modelfit = zeros(numboots,length(xvals)); params = zeros(numboots,2); for boot=1:numboots % prepare data indices ix = ceil(n*rand(1,n)); % construct regressor matrix X = [x(ix)' ones(n,1)]; % estimate parameters h = inv(X'*X)*X'*y(ix)'; % evaluate the model modelfit(boot,:) = xvals*h(1) + h(2); % record the parameters params(boot,:) = h; end % visualize figure; hold on; h1 = scatter(x,y,'k.'); modelfitP = prctile(modelfit,[2.5 50 97.5],1); % define a polygon by following the 2.5th percentile line (left to right) % and then reversing and following the 97.5th percentile line (right to left). h2 = patch([xvals fliplr(xvals)],[modelfitP(1,:) fliplr(modelfitP(3,:))],[1 .7 .7]); set(h2,'EdgeColor','none'); h3 = plot(xvals,modelfitP(2,:),'r-','LineWidth',2); uistack(h1,'top'); % make sure data points are visible by bringing them to the top xlabel('x'); ylabel('y'); paramsP = prctile(params,[2.5 97.5],1); title(sprintf('y=ax+b; 95%% confidence intervals: a=[%.1f %.1f], b=[%.1f %.1f]', ... paramsP(1,1),paramsP(2,1),paramsP(1,2),paramsP(2,2)));
Example 3: Perform leave-one-out cross-validation for a simple linear model
% reuse data from previous example n = n; x = x; y = y; % perfom leave-one-out cross-validation predictions = zeros(1,n); % this will hold the prediction for each data point for p=1:n % figure out indices trainix = setdiff(1:n,p); testix = p; % train the model X = [x(trainix)' ones(length(trainix),1)]; % construct regressor matrix h = inv(X'*X)*X'*y(trainix)'; % estimate parameters % test the model by computing the prediction for the left-out data point predictions(p) = [x(testix)' ones(length(testix),1)]*h; end % quantify how well the predictions match the data using coefficient of determination R2 = 100 * (1 - sum((y-predictions).^2) / sum((y-mean(y)).^2)); % report the answer fprintf('Cross-validated R^2 = %.2f%%\n',R2);
Cross-validated R^2 = 87.89%